UPS and E3s for Geeks!
UPS Primer
All proteins, sooner or later, are naturally degraded by our bodies. Degradation is one means to regulate the amounts of protein or their levels of activity in a cell.
The UPS regulates virtually all cellular processes, such as cell growth, proliferation, migration, signaling, and death. Our targeted protein degrader technology harnesses the body’s innate ubiquitin-proteasome system (UPS) to destroy disease-causing proteins.
Ubiquitin is a small protein present in all organisms and, as the name suggests, is expressed ubiquitously in all cells in our body. Ubiquitin is joined to other proteins (substrates) by a process known as ubiquitylation. Ubiquitylation can conjugate either a single ubiquitin (monoubiquitylation) or create a chain of ubiquitins (polyubiquitylation) on lysine residues of substrate proteins. The polyubiquitin chain functions as an ID tag that marks substrate proteins for the delivery to the proteasome, a large protein complex in which protein degradation takes place.
Tagging substrate proteins with ubiquitin (ubiquitylation), is a highly sophisticated biochemical process catalyzed by a cascade of three enzymes. First, ubiquitin is “activated” by being transferred to a ubiquitin-activating enzyme (E1); this reaction requires the energy-rich molecule ATP. Second, the activated ubiquitin is transferred from E1 to a ubiquitin-conjugating enzyme (E2). Finally, a ubiquitin ligase (E3) transfers the ubiquitin from E2 to a lysine residue in the substrate protein.
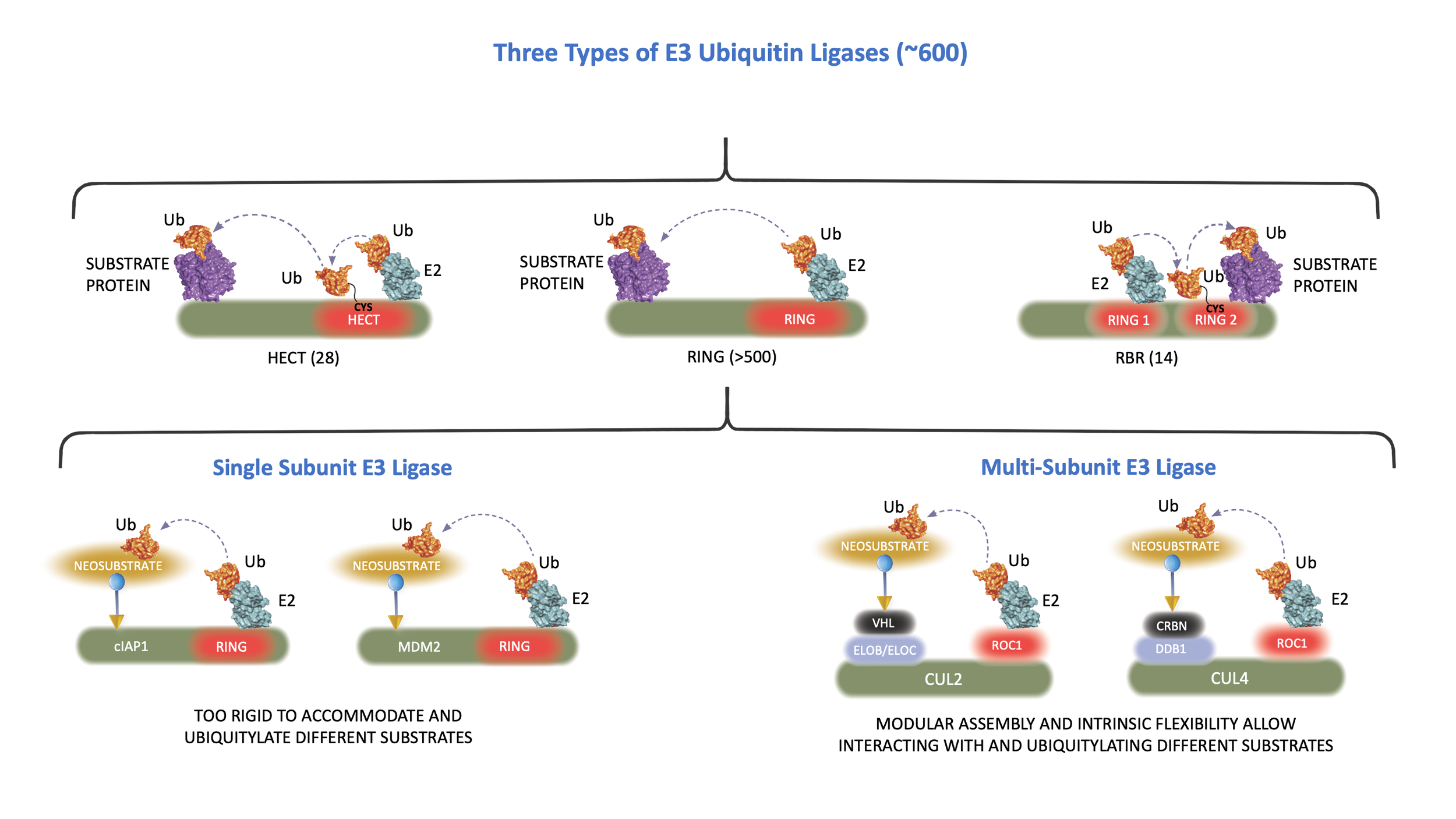
The E3 ligase determines when, where, and which proteins are tagged with ubiquitin. Human cells possess an estimated more than 600 E3 ligases, each specific for particular protein targets. Building upon this precise degradation system and 20 years of research experiences, scientists at Cullgen are exploring novel strategies to target selective disease-causing proteins for the degradation by the UPS
E3 ligases can be used by small molecules to degrade disease-causing proteins
The vast majority of E3 ubiquitin ligases have a structure known as the RING finger. The RING finger mediates ubiquitin transfer from E2 to a target protein. There are two major types of RING finger E ligases: One containing an intrinsic RING finger domain and the other using a scaffold protein, cullin, to bind to a small RING finger protein, ROC1 or ROC2. Individual cullin proteins can bind with multiple different substrate receptor proteins, either directly, such as CUL3, or indirectly through a linker protein, such as ELOB/ELOC in the case of cullin 2 (CUL2) and DDB1 in the case of CUL4. Each cullin can associate with many different substrate receptors, leading to the assembly of an estimated more than 300 distinct cullin-RING E3 ubiquitin ligases (CRLs). The modular assembly of CRL ligase complexes provides the flexibility to specifically target the degradation of a large number of substrate proteins. This flexibility can also be reprogrammed by small molecules to degrade proteins of interest.